Time and again I see people making conclusions between Finnish N1c1 and eastern admixture. Regardless of the eastern origin of N1c there is not such correlation in Finland. The reality is even worse for those who cherish this fallacy; if we count also Baltic countries the correlation turns out to be negative. In Finland alone all male haplogroups have equal level Asian admixture and the only difference comes from the locality, not from the male haplogroup. Rational person would conclude that the Asian admixture is from a local source. This is a no-brainer and I don't even need to prove it. Everyone being familiar with this matter knows it, but it doesn't prevent the biggest Finnish newspaper distributing this urban myth. Google translation, click here.
Epilogue. The fallacy of the eastern origin of Finns results from many things. I am not interested in other opinions than those bothering Finnish people and researchers, because I don't care much about "public opinions" without scientific basis. A common idea in Finland, believing in different Finnish origins (Lappeenranta-Vaasa or whatever axis) driven by Finnish "race realists" who inherited opinions from the old Swedish school, is that here in Finland have lived two "races". Now some Finnish scientists have agreed this and detached themselves from known historic facts.
Saturday, December 29, 2018
Thursday, December 27, 2018
QpAdm - what it means in practice
As we saw in my previous posts the correlation between fit and standard error is very meaningful. We saw that the Basques are a loose mixture of East European Steppe and ancient Iberian people, but they are only far descendants of those two groups and we can't prove that these two are their only ancestor, although they definitely forwarded genes to Basques. I made similar test showing that the Greeks are distant descendants of Iron Age Anatolians and Bronze Age Balkanians, but again we can't be prove that those two were their only ancestors. Probably not.
Balkans_BronzeAge Anatolia_IA
best coefficients: 0.470 0.530
Jackknife mean: 0.475197121 0.524802879
std. errors: 0.077 0.077
fixed pat wt dof chisq tail prob
00 0 8 15.062 0.0579554 0.470 0.530
On the other hand, qpAdm showed that the Finns are very strictly descendants of Iron Age Scanian, Iron Age Baltic and Iron Age Saami people, but we can't prove exact proportions of those thee admixtures, which we saw in high standard errors. It is easy to understand that admixtures of close populations are not as easy determinable as admixtures of distant populations, because close relatives share much common ancestry.
But how accurate are results showing very distant ancestry and moderately low standard errors, if the fit is poor? I tested it. Following tests show admixtures of Iron Age Saami people in Ostrobothnia Levaluhta.
We see that there is only a small difference in admixtures of Iron Age Saamis generated by present-day Finns and Iron Age Scandinavians in conjunction of Bolshoy outlier. Chisq is high, tail prob. below 0.4, but std. err. only 6% max. Nothing obliges such a high admixture similarity, because the genetic distance between Finns and Scandinavians is rather high. Such a similarity is achieved only by a big genetic distant of Bolshoy outlier.
Another example, although not equally striking.
Chisq is between 10 and 21, tail prob. between 0.006 and 0.24. FI21 shows best fit. Std.error is 5% in FI4 and FI12, highest (9%) in case of FI21.
Balkans_BronzeAge Anatolia_IA
best coefficients: 0.470 0.530
Jackknife mean: 0.475197121 0.524802879
std. errors: 0.077 0.077
fixed pat wt dof chisq tail prob
00 0 8 15.062 0.0579554 0.470 0.530
On the other hand, qpAdm showed that the Finns are very strictly descendants of Iron Age Scanian, Iron Age Baltic and Iron Age Saami people, but we can't prove exact proportions of those thee admixtures, which we saw in high standard errors. It is easy to understand that admixtures of close populations are not as easy determinable as admixtures of distant populations, because close relatives share much common ancestry.
But how accurate are results showing very distant ancestry and moderately low standard errors, if the fit is poor? I tested it. Following tests show admixtures of Iron Age Saami people in Ostrobothnia Levaluhta.
We see that there is only a small difference in admixtures of Iron Age Saamis generated by present-day Finns and Iron Age Scandinavians in conjunction of Bolshoy outlier. Chisq is high, tail prob. below 0.4, but std. err. only 6% max. Nothing obliges such a high admixture similarity, because the genetic distance between Finns and Scandinavians is rather high. Such a similarity is achieved only by a big genetic distant of Bolshoy outlier.
Another example, although not equally striking.
Chisq is between 10 and 21, tail prob. between 0.006 and 0.24. FI21 shows best fit. Std.error is 5% in FI4 and FI12, highest (9%) in case of FI21.
Saturday, December 22, 2018
Exciting results of Basques and Estonians, updated: Southwest Finnish results
I made recalibration of qpAdm references to improve accuracy. Please read my previous post to find my opinion and to see the problematic with qpAdm.
New references:
Kostenki14 813405
MA1 625746
WHG 703903
EHG 975726
CHG 889688
Ganj_Dareh_N 794892
West_Siberia_N 670626
Anatolia_Neolithic 889986
Mbuti1M 971767
Wichi1M 971774
At first I tried to find out the admixture of present-day Estonians and it is really challenging without proper Iron Age samples. So I had to use best available modern samples.
Latvian1M FI12 AncFinn
best coefficients: 0.408 0.451 0.141
Jackknife mean: 0.404965620 0.441429933 0.153604447
std. errors: 0.248 0.159 0.203
fixed pat wt dof chisq tail prob
000 0 7 2.724 0.90932 0.408 0.451 0.141
001 1 8 3.392 0.907406 0.531 0.469 0.000
010 1 8 10.133 0.255821 2.398 -0.000 -1.398 infeasible
100 1 8 5.005 0.757038 0.000 0.626 0.374
Admixtures:
Latvian1M - Latvian samples covering 1 million SNP's
FI12 - it is me, because I am one of my individual samples covering 1 million SNP's and in this case giving best fit. So I represent here present-day Finns.
AncFinn - an ancient Finnish sample from Damgaard et al. 2018.
Reasonable fit for Basques was even more challenging. My test shows that the Basques are averagely two thirds ancient people from the Iberian peninsula and one third from Steppe origin.
SE_Iberia_CA Yamnaya_Samara
best coefficients: 0.671 0.329
Jackknife mean: 0.670091689 0.329908311
std. errors: 0.022 0.022
fixed pat wt dof chisq tail prob
00 0 8 5.956 0.652145 0.671 0.329
edit 25.12.2018 12:55
A new Southwest Finnish result using recalibrated references. Recalibration here means better results of Asian and African admixtures. There was also a inconsistency between Iron Gate and WHG - Iron Gate removed.
Scania_IA Baltic_IA Levaluhta
best coefficients: 0.483 0.358 0.159
Jackknife mean: 0.450272111 0.373492239 0.176235650
std. errors: 0.182 0.183 0.111
fixed pat wt dof chisq tail prob
000 0 7 0.657 0.998647 0.483 0.358 0.159
001 1 8 2.229 0.97317 0.596 0.404 0.000
010 1 8 3.018 0.93322 0.803 0.000 0.197
100 1 8 5.884 0.660177 -0.000 0.849 0.151
011 2 9 4.219 0.896439 1.000 0.000 0.000
101 2 9 6.568 0.681978 0.000 1.000 0.000
110 2 9 21.991 0.00890644 0.000 0.000 1.000
New references:
Kostenki14 813405
MA1 625746
WHG 703903
EHG 975726
CHG 889688
Ganj_Dareh_N 794892
West_Siberia_N 670626
Anatolia_Neolithic 889986
Mbuti1M 971767
Wichi1M 971774
At first I tried to find out the admixture of present-day Estonians and it is really challenging without proper Iron Age samples. So I had to use best available modern samples.
Latvian1M FI12 AncFinn
best coefficients: 0.408 0.451 0.141
Jackknife mean: 0.404965620 0.441429933 0.153604447
std. errors: 0.248 0.159 0.203
fixed pat wt dof chisq tail prob
000 0 7 2.724 0.90932 0.408 0.451 0.141
001 1 8 3.392 0.907406 0.531 0.469 0.000
010 1 8 10.133 0.255821 2.398 -0.000 -1.398 infeasible
100 1 8 5.005 0.757038 0.000 0.626 0.374
Admixtures:
Latvian1M - Latvian samples covering 1 million SNP's
FI12 - it is me, because I am one of my individual samples covering 1 million SNP's and in this case giving best fit. So I represent here present-day Finns.
AncFinn - an ancient Finnish sample from Damgaard et al. 2018.
Reasonable fit for Basques was even more challenging. My test shows that the Basques are averagely two thirds ancient people from the Iberian peninsula and one third from Steppe origin.
SE_Iberia_CA Yamnaya_Samara
best coefficients: 0.671 0.329
Jackknife mean: 0.670091689 0.329908311
std. errors: 0.022 0.022
fixed pat wt dof chisq tail prob
00 0 8 5.956 0.652145 0.671 0.329
edit 25.12.2018 12:55
A new Southwest Finnish result using recalibrated references. Recalibration here means better results of Asian and African admixtures. There was also a inconsistency between Iron Gate and WHG - Iron Gate removed.
Scania_IA Baltic_IA Levaluhta
best coefficients: 0.483 0.358 0.159
Jackknife mean: 0.450272111 0.373492239 0.176235650
std. errors: 0.182 0.183 0.111
fixed pat wt dof chisq tail prob
000 0 7 0.657 0.998647 0.483 0.358 0.159
001 1 8 2.229 0.97317 0.596 0.404 0.000
010 1 8 3.018 0.93322 0.803 0.000 0.197
100 1 8 5.884 0.660177 -0.000 0.849 0.151
011 2 9 4.219 0.896439 1.000 0.000 0.000
101 2 9 6.568 0.681978 0.000 1.000 0.000
110 2 9 21.991 0.00890644 0.000 0.000 1.000
Tuesday, December 18, 2018
Still not enough West European Iron Age samples to get proper qpAdm results of West Europeans
My try to model present-day Swedes was not what I hoped, because lack of proper western Iron Age samples. Now I tried to find out the best possible solution using Scania_IA and older samples. I noticed that in all possible variations we need recently unavailable and unknown Iron Age samples to achieve reasonable results. So I have to forget such tests until West European Iron Age samples are available. Several Central European Late Copper Age samples turned out to be best ones, but made not proper fits, for instance:
Scania_IA Protoboleraz_LCA
best coefficients: 0.949 0.051
Jackknife mean: 0.947619305 0.052380695
std. errors: 0.041 0.041
This is best I can do right now.
An issue beyond qpAdm is how to determine standard errors. While we can consider low standard error good, there is also a good reason to consider high standard error reasonable in many cases. In a case where two or more populations share pretty much common ancestry (as it is in many case today) qpAdm can't determine which one is the right one. For instance in a case of admixtures built of Swedes and Norwegians the standard error can be very high, because qpAdm is not able to break ancestries into common ancestry of both populations. So, when we try to minimize the standard error we in fact abandon the most obvious result. Usually this dilemma is tried to avoid in two ways: 1) using very ancient/distant samples to avoid common ancestry or 2) approving very high chisq and small tail prob values. In the latter case we actually approve poorer results to show falsely better results.
A result showing high standard errors:
Estonians:
Scania_IA Baltic_IA Poland_BA
best coefficients: 0.560 0.108 0.332
Jackknife mean: 0.253950408 0.349222728 0.396826864
std. errors: 0.532 0.634 0.389
In this case all admixtures are overlapping resulting statistical transitions and uncertainty between admixtures and high standard errors, but chisq and tail prob values are still relatively good, respectively 2.290 and 0.942093.
Another case shows low standard errors, but poorer coverage of admixtures:
Swedes:
Scania_IA Hungary_LCA
best coefficients: 0.948 0.052
Jackknife mean: 0.946235880 0.053764120
std. errors: 0.043 0.043
Respectively chisq and tail prob values were 7.413 and 0.492767.
I can make a more provocative latter example for similar target populations in which standard errors could be 1-2 percentages and chisq and tail prob values around 10-20 and 0.1-0.2
Scania_IA Protoboleraz_LCA
best coefficients: 0.949 0.051
Jackknife mean: 0.947619305 0.052380695
std. errors: 0.041 0.041
This is best I can do right now.
An issue beyond qpAdm is how to determine standard errors. While we can consider low standard error good, there is also a good reason to consider high standard error reasonable in many cases. In a case where two or more populations share pretty much common ancestry (as it is in many case today) qpAdm can't determine which one is the right one. For instance in a case of admixtures built of Swedes and Norwegians the standard error can be very high, because qpAdm is not able to break ancestries into common ancestry of both populations. So, when we try to minimize the standard error we in fact abandon the most obvious result. Usually this dilemma is tried to avoid in two ways: 1) using very ancient/distant samples to avoid common ancestry or 2) approving very high chisq and small tail prob values. In the latter case we actually approve poorer results to show falsely better results.
A result showing high standard errors:
Estonians:
Scania_IA Baltic_IA Poland_BA
best coefficients: 0.560 0.108 0.332
Jackknife mean: 0.253950408 0.349222728 0.396826864
std. errors: 0.532 0.634 0.389
In this case all admixtures are overlapping resulting statistical transitions and uncertainty between admixtures and high standard errors, but chisq and tail prob values are still relatively good, respectively 2.290 and 0.942093.
Another case shows low standard errors, but poorer coverage of admixtures:
Swedes:
Scania_IA Hungary_LCA
best coefficients: 0.948 0.052
Jackknife mean: 0.946235880 0.053764120
std. errors: 0.043 0.043
Respectively chisq and tail prob values were 7.413 and 0.492767.
I can make a more provocative latter example for similar target populations in which standard errors could be 1-2 percentages and chisq and tail prob values around 10-20 and 0.1-0.2
Monday, December 10, 2018
Finnish genetic composition: Iron Age Baltic, Iron Age Germanic and probably Iron Age Saami people
You probably have read my previous post regarding European genetic structures composed by Admixture and Eurasian data. My aim was to make an admixture analysis free of recent genetic drift. It gave following admixtures for Southwestern Finns (Finnish, k=10)
- Saami 14%
- Baltic 49%
- Germanic 35%
The post is here.
Now I have tested same admixtures using Iron Age samples and qpAdm. QpAdm allocates admixtures for given populations, in these tests admixture populations were Baltic Iron Age, Scanian Iron Age and Levaluhta Iron Age. Levaluhta consists of five Iron Age remains found from Finnish Bothnia, Ostrobothnia. The id of the Scanian IA sample is RISE174 and the Baltic IA sample is DA171. Although my results are unambiguous, there are some uncertainty regarding the software and given references, and being the first exploring something like this I am curious to see results of professional geneticists. I simply can't understand why this matter wouldn't interest also researchers. All samples I use here are publicly available.
Southwest Finns:
chisq tail prob
1.112 0.992818
Levaluhta 0.115
Scania_IA 0.437
Baltic_IA 0.448
Vepsa:
chisq tail prob
1.555 0.980354
Levaluhta 0.202
FIN_Southwest 0.614
Baltic_IA 0.184
Nganasan -0.000
and another result of Vepsa
chisq tail prob
2.734 0.908488
Levaluhta 0.000
FIN_Southwest 0.764
Baltic_IA 0.164
Nganasan 0.072
The match is much worse when using present-day Baltic, Germanic and Saami counterparts.
Southwest Finns:
chisq tail prob
9.270 0.233881
Saami 0.156
Latvian 0.297
Swedish 0.548
Outgroups and coverages were
Kostenki14 813405
MA1 625746
WHG 703903
Iron_Gates_HG 884901
EHG 983976
CHG 889688
Ganj_Dareh_N 794892
West_Siberia_N 670626
Anatolia_Neolithic 889986
LBK_EN 880957
Typical SNP coverage of target groups
Vepsa1M 971774
Levaluhta 377788
FIN-Southwest 1102712
Baltic_IA 97311
Scania_IA 373352
Nganasan1M 971774
I have lost some SNP's due to allele mismatches or multiallelic conversion errors in Plink, but the coverage is still reasonable. Some ancient samples need to be reconverted.
edit 11.12.2018 16:30
Adding Russians from Pinega makes a perfect match for Vepsas. Not a big surprise.
chisq tail prob
0.825 0.991377
FIN-Southwest 0.672
RusPinega 0.130
Baltic_IA 0.093
Levaluhta 0.105
- Saami 14%
- Baltic 49%
- Germanic 35%
The post is here.
Now I have tested same admixtures using Iron Age samples and qpAdm. QpAdm allocates admixtures for given populations, in these tests admixture populations were Baltic Iron Age, Scanian Iron Age and Levaluhta Iron Age. Levaluhta consists of five Iron Age remains found from Finnish Bothnia, Ostrobothnia. The id of the Scanian IA sample is RISE174 and the Baltic IA sample is DA171. Although my results are unambiguous, there are some uncertainty regarding the software and given references, and being the first exploring something like this I am curious to see results of professional geneticists. I simply can't understand why this matter wouldn't interest also researchers. All samples I use here are publicly available.
Southwest Finns:
chisq tail prob
1.112 0.992818
Levaluhta 0.115
Scania_IA 0.437
Baltic_IA 0.448
Vepsa:
chisq tail prob
1.555 0.980354
Levaluhta 0.202
FIN_Southwest 0.614
Baltic_IA 0.184
Nganasan -0.000
and another result of Vepsa
chisq tail prob
2.734 0.908488
Levaluhta 0.000
FIN_Southwest 0.764
Baltic_IA 0.164
Nganasan 0.072
The match is much worse when using present-day Baltic, Germanic and Saami counterparts.
Southwest Finns:
chisq tail prob
9.270 0.233881
Saami 0.156
Latvian 0.297
Swedish 0.548
Outgroups and coverages were
Kostenki14 813405
MA1 625746
WHG 703903
Iron_Gates_HG 884901
EHG 983976
CHG 889688
Ganj_Dareh_N 794892
West_Siberia_N 670626
Anatolia_Neolithic 889986
LBK_EN 880957
Typical SNP coverage of target groups
Vepsa1M 971774
Levaluhta 377788
FIN-Southwest 1102712
Baltic_IA 97311
Scania_IA 373352
Nganasan1M 971774
I have lost some SNP's due to allele mismatches or multiallelic conversion errors in Plink, but the coverage is still reasonable. Some ancient samples need to be reconverted.
edit 11.12.2018 16:30
Adding Russians from Pinega makes a perfect match for Vepsas. Not a big surprise.
chisq tail prob
0.825 0.991377
FIN-Southwest 0.672
RusPinega 0.130
Baltic_IA 0.093
Levaluhta 0.105
Tuesday, December 4, 2018
Finnish and North Russian ancestries derived from Levaluhta and Bolshoy Oleni Ostrov
Unfortunately the Lamnidis et al. 2018 did not test the origin of the Siberian ancestry in present-day Finns and Russians. I reveal it now. Ancient samples found from Bolshoy gives high F3-results for both groups. I included five Finnish groups, three of my own sample selection and two smaller selections from two academic sources.
FinnMostCW - least drifted Finnish group (- outliers)
FinnLocal - most drifted Finnish group (- outliers)
Finn21M - most Scandinavian group (- outliers)
Both academic Finnish groups look incoherent. I wonder why.
It is obvious that the Levaluhta admixture added to the present-day European backbone in Finns (negative F3-values) doesn't explain all non-Europeanness and there have to be an unknown third component.
Source1, source2, target, f_3, std.err, Z and SNP count
Additionally, here are resultw of my old admixture analysis based on Dna.Land's program. Although it sounds a weird idea to make admixture analysis of ancient samples using present-day populations, it is a normal practice in many academic studies. Studies usually use Nganasans to represent Siberians together with ancient samples. It is reasonable to say that the gene flow can be detected also backwards. It is also very likely that the gene flow between contemporary populations has been bidirectional. Nevertheless, I am not fully convinced about the gene flow direction regarding the European side of Finns and Northern Russians. F3-statistics gave several European candidates and the big picture is more complex.
Levaluhta (JK excluding JK2065 outlier)
Finnic 43,7
Saami 30,0
Uralic 12,1
Siberian 9,5
Baltic 2,2
East_Asian 1,9
Bolshoy
Uralic 38.9
Siberian 29.5
Saami 11.6
Finnic 7.1
East_Asian 6.1
Northeast_European 5.4
Baltic 1.5
Wednesday, November 28, 2018
New study tells about the Siberian origin, but not yet about the Finns
Finally, the new study about the Siberian origin in Finland is published. It is however too early to say where the Finns came from, or even speculate on it, because the study includes no samples from the settlement areas of ancient (prehistoric) Finns or from areas where they according scientists came from. Rumors tell that researchers have Iron Age samples also from the Southwest Finland, but we have still to wait to see it. The data is not yet available, but I'll be online soon after I have them.
Sunday, November 25, 2018
Perspectives using traditional unsupervised admixture analysis
Trying to border us closer on our reality and on the side of believable I have done the following series of Admixture analyses using samples of living Eurasian populations. My database consists of 0.5 million SNP's over the world, but to minimize the effect of recent genetic drift I made a LD pruning operation decreasing the amount of SNP's to 125 kSNP's. Without that many isolated sample groups would hijack the analysis and turn the history backward. But there are many kinds of genetically isolated populations; the Basques are not a young isolation. Conversely in large areas in the North Europe and in the North Asia many populations are very young and they can make the history really backward in genetic tests. Those small northern, not so long ago diverged groups, were hunter-gatherers or nomads without borders. Once western or eastern civilizations with text books appeared to the neighborhood they were localized and they we named. This happened only a few hundreds years ago. No one really know where they lived and where they came from THOUSANDS years ago.
This post just to remind about the present-day reality after hypocrite uniparental speculations about our ancestry seen on the internet and in popular science. The command line was
$admixture1.3 data.bed n -j4 -B
where n is 9-15 and 20 (20 only in numeric data) .
You can repeat this test with your own data and post results here. Remember to carry out LD-pruning to get rid of the misleading recent genetic drift.
The data includes 560 individual, 20 samples at its peak in each group. I computed group averages to make the results more easy to read. The data behind bars can be downloaded here.
Be aware of the bar colors, because I had not nerve to arrange colors manually and automated "R barplot" coloring was not a bullet proof solution and same colors exist repeatedly with several k values. You can check the result using downloaded data.
Tuesday, November 6, 2018
Mongolian or not Mongolian
I already promised to stop my IBD rant, but it did happen that a new Chinese study provoked me to do a little more. This new study is here and here. The figure below shows shared IBD segments of 1-2 cM and the ratio between shared segments of Evenks and Mongolians. If the ratio is high then the Evenk admixture is more likely than the Mongolian one. If the ratio is low, but over 1, we go closer the Mongolian probability, but the Evenks IBD (Siberians) can act like a proxy for Mongolians. Ratios below 1 means that the Mongolian admixture is more likely than the Evenk (Siberian) one.
Friday, November 2, 2018
Pskov Russia, the connection between Finnic groups
This will be my last IBD test for a while, because IBD itself is not a good method for seeking oldest ancestral linkages. It is still a good statistic method manifesting events of the Iron Age and the genealogical time frame. It is excellent in seeking real ancestral linkages in the before mentioned time-period and can be preferred instead of any method using allele data, such as PCA, DSTAT etc., because all those methods imply only genetic distance and genetic distance depends on the admixture itself. Using allele statistic in seeking the origin of mixed populations is always misleading. You can only suggest that your hypotheses are right or wrong and continue testing using different variations to increase the probability.
Here I present some IBD results of both Finnic and Russian populations using Beagle version 5. The linkage between all Finnic people and also between Finnic and Baltic people is notable and turns up in Pskov (in Finnish Pihkova). The Pskov area is also the northernmost place in Russia where I have found autosomal relatives (23andme).
I use again Finnish samples from the 1000 genomes project, but at this time I grouped them using my project members, whose geographic origin is known.
Cross-checking with Pskov is presented below. Notice that there is no Siperian admixture in Pskov. Although people, including most researches, suggest that the Siberian admixture among Finnic people is a common signature of the origin of the FU languages, nothing about it is proven. The Siberian admixture in Finland is dated to the Iron Age by several scientists and research. Still the hypothesis of the present day Siberian admixture in Finland and the origin of the language (FU urheimat theories) is usually thought to be proven! The IBD connection between Finns and North Russian is two-fold; at first the Finnish Iron Age migration to Russia and secondly the common Siberian admixture which is quite new. This kind of Iron Age Siberian migration/admixture doesn't exist between Finns and people in Pskov and we have no evidence about migrations from Finland to Pskov. Instead of this, we have clear and undeniable linguistic evidences about Finnish eastern migrations to the Northwestern Russia, at least to the area of the lake Onega! The primary conclusion have to be that the Pskov area had a pre-Iron Age connection with Finnic people, including the Finns. It must have happened before the Siberian admixture in Finland and before the Finnish eastern migration.
Russian results do not give an observation of any distinct ancestry from the Pskov area and only the northernmost Russians show some divergence from the Russian resemblance.
Here I present some IBD results of both Finnic and Russian populations using Beagle version 5. The linkage between all Finnic people and also between Finnic and Baltic people is notable and turns up in Pskov (in Finnish Pihkova). The Pskov area is also the northernmost place in Russia where I have found autosomal relatives (23andme).
I use again Finnish samples from the 1000 genomes project, but at this time I grouped them using my project members, whose geographic origin is known.
Cross-checking with Pskov is presented below. Notice that there is no Siperian admixture in Pskov. Although people, including most researches, suggest that the Siberian admixture among Finnic people is a common signature of the origin of the FU languages, nothing about it is proven. The Siberian admixture in Finland is dated to the Iron Age by several scientists and research. Still the hypothesis of the present day Siberian admixture in Finland and the origin of the language (FU urheimat theories) is usually thought to be proven! The IBD connection between Finns and North Russian is two-fold; at first the Finnish Iron Age migration to Russia and secondly the common Siberian admixture which is quite new. This kind of Iron Age Siberian migration/admixture doesn't exist between Finns and people in Pskov and we have no evidence about migrations from Finland to Pskov. Instead of this, we have clear and undeniable linguistic evidences about Finnish eastern migrations to the Northwestern Russia, at least to the area of the lake Onega! The primary conclusion have to be that the Pskov area had a pre-Iron Age connection with Finnic people, including the Finns. It must have happened before the Siberian admixture in Finland and before the Finnish eastern migration.
Russian results do not give an observation of any distinct ancestry from the Pskov area and only the northernmost Russians show some divergence from the Russian resemblance.
Wednesday, October 10, 2018
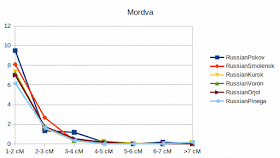
More IBD-statistics, including Mordvas, Brits and Basques
I have now automated the process and can do "quick and dirty" statistics of all below listed populations. Adding new populations takes about two days in a case I have them in my backbone data base. Here are now existing populations and sample counts. Adding new populations to my data base takes longer, around a week.
The results are based on slightly changed phasing parameters, so there can be slight differences in absolute values. Used programs are Plink, Impute2 and Beagle version 5.
The results are based on slightly changed phasing parameters, so there can be slight differences in absolute values. Used programs are Plink, Impute2 and Beagle version 5.
Sunday, September 30, 2018
Potential Kyrgyz admixture in Europe, shown using IBD results from Beagle 5
It is well known that during the first millennium the Europe was threatened by invading Mongols called Huns. The Huns themselves were probably not a homogeneous group and were build of many ethnic groups. Later, during the second millennium Europeans, especially Eastern Slavs were threatened by the army of Genghis Khan. Those later rulers were called Tatars. My results show that there is a subtle Mongol admixture in Europe, but amazingly not among present day Tatars. The results are based on the difference of IBD sharing between Nganasans and Kyrgyz. Nganasans are known as an isolated group of Northwest Siberian people, used often to demonstrate Siberian admixture in Europe. My previous test shows that Evens match even better in this purpose, but the distribution of Evens is much larger in Europe, which can be due to the European admixture among Evens, rather than wise versa. So it is better to use more distinct Nganasans in this purpose.
Saturday, September 29, 2018
Swedish IBD-sharings around the Baltic Sea and a little further
Unfortunately I have not enough German, Danish and Norwegian samples, or the origin is unclear, but here some evidence about the Swedish influence in Finland, or conversely Finnish/Karelian influence in Sweden, if you prefer it. For better or worse, the IBD-sharing is evident on the eastward route. My goal is to add at least some known German samples in the future.
Saami IBD-sharing
Continuing with the same phased data I made two IBD-statistics and compared the Saami (from Finnmark) with Northern Europeans and Siberians. The result shows highest 1-2 cM IBD's between Saamis and Even samples from the SGDP data. So I made a follow-up using SGDP-Evens. I have not detailed information about the origin of those Evens. The curve implies that the connection between Saamis and SGDP-Evens is old but strong.
Notable is also the strong 2-3cM sharing between Saamis and Northern Baltic Finns, including Finns, Karelians, Vepsas and Ingrians. This may be a consequence of the first contact between Saamis and Baltic Finns. Smaller segments are more vague in timing due to the random segment break down and data inaccuracy.
Notable is also the strong 2-3cM sharing between Saamis and Northern Baltic Finns, including Finns, Karelians, Vepsas and Ingrians. This may be a consequence of the first contact between Saamis and Baltic Finns. Smaller segments are more vague in timing due to the random segment break down and data inaccuracy.
Friday, September 28, 2018
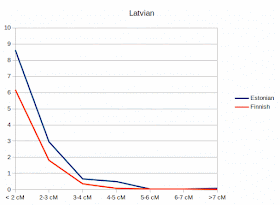
Differences in IBD-sharings between Estonians and Finns
These statistics are based on haploid IBD's made by Beagle 5 and Beagle Refined IBD. The data was partly imputed (IMPUTE2) to increase the SNP amount of some populations from 300 kSNP to 500kSNP, although most samples are from larger data sets. Samples of this IBD-data are listed here, including also sample sizes. Accuracy of results depends on the sample size, although trends are quite reliable anyway, as the following tests show without doubt. I'll continue testing with this data later.
Results show the average IBD-sharing (segments) between single individuals of each population.
(X-axis legends fixed, note that the minimum segment size was 1 cM)
Results show the average IBD-sharing (segments) between single individuals of each population.
(X-axis legends fixed, note that the minimum segment size was 1 cM)
Sunday, September 9, 2018
Sigtuna, Sweden Viking Age
I was lucky to get new samples (ENA bam-files) from the study Genomic and Strontium Isotope Variation Reveal Immigration Patterns in a Viking Age Town, Maja Krzewiska et al. 2018. The study included seven high quality late Viking Age samples from the Swedish town Sigtuna. Sigtuna was an important market place, founded 970 ad and continued to be important to the 13th century. These seven samples are good enough to make admixture analyses. I have made admixture analyses using Dna.Land's software, look here, and I see no reason to change my methods. These samples are only 900-1100 years old and it is reasonable to suggest that modern references are suitable for this purpose, especially because with the modern reference the genome quality is is much better than using other ancient samples as references.
Why admixture analyses, why not tests utilizing genetic drift, like qp3Pop and qpDstat? Both above-mentioned methods or other tests like IBD and IBS statistics, as well as Fst tell genetic distances, not the genome structure, giving often a false image of our ancestry. In spite of weaknesses of admixture analyses in case of really old ancient samples (where no one can really figure the admixture history), in this particular case all samples are only 1000 years old and the link between them and us is significant and the admixture history figurable. Good admixture results only call for two condition to be true: at first the admixture history must be real and references must be right. In many cases these conditions are not fulfilled and people after seeing senselessness start to bark up the wrong tree.
grt036
East_Scandinavian 24.7
Slavic 18.8
Mediterranean 14.3
Northeast_European 14.0
Central_European 11.0
Northwest_European 8.8
Saami 4.4
Baltic 3.3
grt035
Northwest_European 40.8
Central_European 16.7
Mediterranean 14.8
East_Scandinavian 10.8
Saami 9.8
Baltic 5.7
Slavic 1.3
kal006 (this looks like present-day Estonians)
Baltic 54.3
Finnic 20.4
East_Scandinavian 12.8
Northeast_European 9.5
Siberian 2.0
stg021 (this looks like preset-day Swedes)
East_Scandinavian 43.5
Northwest_European 22.9
Mediterranean 12.8
Slavic 6.6
Saami 6.1
Northeast_European 2.5
Uralic 1.3
East_Asian 1.8
Baltic 1.7
84001 (this must be mainly British)
Northwest_European 64.2
Baltic 14.1
Uralic 5.0
Saami 4.7
East_Scandinavian 4.3
Slavic 2.1
Mediterranean 2.3
Finnic 2.8
84005 (a quarter Finn, maybe even more Finnish taking into account the Finnish demographic history after the Viking Age)
East_Scandinavian 39.6
Finnic 24.7
Baltic 13.6
Northwest_European 11.8
Slavic 3.1
Mediterranean 3.0
Central_European 3.6
For a comparison, a project sample of Finnish ancestry from Southwestern Finland:
Finnic 49.1
Northwest_European 25.1
East_Scandinavian 12.9
Northeast_European 5.1
Slavic 2.4
Saami 2.9
Baltic 2.3
urm160
Central_European 33.1
East_Scandinavian 29.9
Slavic 19.1
Finnic 6.8
Baltic 4.5
Uralic 2.9
Saami 1.8
Northwest_European 1.3
It is notable that many Viking Age Swedish samples show Saami without Finnish ancestry. This probably means that the Finnish-Saami mixing was not yet as significant as today. It is also possible that the Saami admixture here means common Swedish-Saami ancestry which is not clearly assignable using modern Saami references.
Why admixture analyses, why not tests utilizing genetic drift, like qp3Pop and qpDstat? Both above-mentioned methods or other tests like IBD and IBS statistics, as well as Fst tell genetic distances, not the genome structure, giving often a false image of our ancestry. In spite of weaknesses of admixture analyses in case of really old ancient samples (where no one can really figure the admixture history), in this particular case all samples are only 1000 years old and the link between them and us is significant and the admixture history figurable. Good admixture results only call for two condition to be true: at first the admixture history must be real and references must be right. In many cases these conditions are not fulfilled and people after seeing senselessness start to bark up the wrong tree.
grt036
East_Scandinavian 24.7
Slavic 18.8
Mediterranean 14.3
Northeast_European 14.0
Central_European 11.0
Northwest_European 8.8
Saami 4.4
Baltic 3.3
grt035
Northwest_European 40.8
Central_European 16.7
Mediterranean 14.8
East_Scandinavian 10.8
Saami 9.8
Baltic 5.7
Slavic 1.3
kal006 (this looks like present-day Estonians)
Baltic 54.3
Finnic 20.4
East_Scandinavian 12.8
Northeast_European 9.5
Siberian 2.0
stg021 (this looks like preset-day Swedes)
East_Scandinavian 43.5
Northwest_European 22.9
Mediterranean 12.8
Slavic 6.6
Saami 6.1
Northeast_European 2.5
Uralic 1.3
East_Asian 1.8
Baltic 1.7
84001 (this must be mainly British)
Northwest_European 64.2
Baltic 14.1
Uralic 5.0
Saami 4.7
East_Scandinavian 4.3
Slavic 2.1
Mediterranean 2.3
Finnic 2.8
84005 (a quarter Finn, maybe even more Finnish taking into account the Finnish demographic history after the Viking Age)
East_Scandinavian 39.6
Finnic 24.7
Baltic 13.6
Northwest_European 11.8
Slavic 3.1
Mediterranean 3.0
Central_European 3.6
For a comparison, a project sample of Finnish ancestry from Southwestern Finland:
Finnic 49.1
Northwest_European 25.1
East_Scandinavian 12.9
Northeast_European 5.1
Slavic 2.4
Saami 2.9
Baltic 2.3
urm160
Central_European 33.1
East_Scandinavian 29.9
Slavic 19.1
Finnic 6.8
Baltic 4.5
Uralic 2.9
Saami 1.8
Northwest_European 1.3
It is notable that many Viking Age Swedish samples show Saami without Finnish ancestry. This probably means that the Finnish-Saami mixing was not yet as significant as today. It is also possible that the Saami admixture here means common Swedish-Saami ancestry which is not clearly assignable using modern Saami references.
Saturday, June 9, 2018
QpAdm tests, Iron Age Scanian sample RISE174
I've been a long time waiting for ancient Finnish samples and it starts to look unlikely that I'll ever see them because the Finnish soil is acid and destroys organic material in one millennium. But don't ever lose hope, it was available all the time from Internet. An old Finnish sample has been available already three years in open data bases. It was found from the Southern Sweden and was labeled as RISE174, dated 427-611 AD, later Scania_IA. That is right, we have had three years an ancient Fennoscandinavian sample, common for all Fennoscandinavians and also for many Baltic people, but obviously because it was found from Southern Sweden it's real value was not noticed before. The reason could also be partly a common prejudice. I wouldn't be much surprised if Finnish researchers in future will find similar Iron Age samples from Southwestern Finland, if it is possible due to the Finnish soil. I'll show that present-day Swedes are not much more related to this sample than present-day Finns. Generally speaking, all qpAdm-results listed below are reliable, although there were many neighboring cultures in the past, being also genetically very similar and in any tests all populations close enough can substitute each other.
QpAdm results are made using distant ancestral references (the right-file) to get best possible coverage. While these results are very reasonable, it is possible to fine-tune all results by using carefully selected less distant references. So these results are directional.
All Finnish groups are from the 1000-genomes project and cover full 1MSNP's, as well as all other groups on the second and third plots. I have seen in my work that testing ancient and modern samples calls for equal coverage of all modern sample groups.
What kind of Fennoscandinavian was the RISE174/Scania_IA? Definitely she was not like a present-day Swede, we can see it in following f3-result below and on PCA-plots.
Next here is a simple PCA-plot, somewhat imperfect one and flat due to a high amount of Asian influence and too few European samples. This plot shows a new Finnish group, Finnx, and binds it to the Finnish ancestry. Finnx forms an ordinary Finnish group with less present-day Swedish admixture than typical Southwest Finns, but also less genetic drift than typical East Finns.
Another plot (vectors 1/2 and 1/3), removed almost all East Asian and Siberian. There is still a touch East Asian forwarded by South and Central Asians. It was necessary to remove East and Siberian components to ignore later Siberian admixture in Finland and see the possible Finnx-Scania_IA connection also on PCA. On the second plot (vectors 1/3) the East Asian is at its least and Scania_IA gives the best match with Finnx.
F3-test showing common genetic drift with Scania_IA:
First some European groups in a demonstrative purpose.
South Italians
Chisq and tail prob: 0.436 0.932651
Populations: Beaker_Central_Europe, Iran_LN, Levant_BA
best coefficients: 0.299 0.169 0.531
Jackknife mean: 0.299685849 0.169473438 0.530840713
std. errors: 0.028 0.032 0.033
North Italians
Chisq and tail prob: 1.738 0.628609
Populations: Beaker_Central_Europe, Levant_BA, Iran_LN
best coefficients: 0.542 0.416 0.042
Jackknife mean: 0.542422483 0.415568791 0.042008725
std. errors: 0.019 0.020 0.020
Basques
Chisq and tail prob: 1.789 0.774528
Populations: Beaker_Central_Europe, Iberia_EN
best coefficients: 0.578 0.422
Jackknife mean: 0.579234989 0.420765011
std. errors: 0.030 0.030
Latvians
Chisq and tail prob: 1.101 0.77684
Populations: Scania_IA, Latvia_MN, Evenk
best coefficients: 0.920 0.092 -0.012
Jackknife mean: 0.918423990 0.093588254 -0.012012243
std. errors: 0.042 0.042 0.016
Poles
Chisq and tail prob: 1.699 0.790857
Populations: Poltavka, Germany_MN
best coefficients: 0.502 0.498
Jackknife mean: 0.502425062 0.497574938
std. errors: 0.039 0.039
Estonians
Chisq and tail prob: 0.642 0.886697
Populations: Scania_IA, Latvia_MN, Evenk
best coefficients: 0.933 0.052 0.015
Jackknife mean: 0.931374297 0.053427705 0.015197998
std. errors: 0.039 0.038 0.015
Mordvas
Chisq and tail prob: 0.503 0.973198
Populations: Sarmatian, Scania_IA
best coefficients: 0.472 0.528
Jackknife mean: 0.475543261 0.524456739
std. errors: 0.079 0.079
Swedes
Chisq and tail prob: 1.003 0.9093
Populations: Latvia_LN, England_N
best coefficients: 0.523 0.477
Jackknife mean: 0.520472837 0.479527163
std. errors: 0.052 0.052
Finnx
Chisq and tail prob: 1.647 0.648853
Populations: Scania_IA, Latvia_MN, Evenk
best coefficients: 0.851 0.084 0.065
Jackknife mean: 0.849474735 0.085348593 0.065176672
std. errors: 0.037 0.036 0.014
What if Mordvas are Polish
Chisq and tail prob: 11.941 0.0177937
Populations: Poltavka, Germany_MN
best coefficients: 0.727 0.273
Jackknife mean: 0.724786064 0.275213936
std. errors: 0.047 0.047
What if Latvians are Polish
Chisq and tail prob: 9.038 0.0601576
Populations: Poltavka, Germany_MN
best coefficients: 0.563 0.437
Jackknife mean: 0.562264229 0.437735771
std. errors: 0.049 0.049
What if Finnx's are Mordva
Chisq and tail prob: 4.728 0.316351
Populations: Sarmatian, Scania_IA
best coefficients: 0.369 0.631
Jackknife mean: 0.377367376 0.622632624
std. errors: 0.093 0.093
What if East Finns are Mordvas
Chisq and tail prob: 6.593 0.159
Populations: Sarmatian, Scania_IA
best coefficients: 0.317 0.683
Jackknife mean: 0.329388055 0.670611945
std. errors: 0.118 0.118
What if Swedes are Baltic
Chisq and tail prob: 1.867 0.867159
Population: Scania_IA, Latvia_MN, Evenk
best coefficients, Jackknife optimisation is negative, 1.000 0.000 -0.000
The results above show the continuum of Scania_IA from Southern Sweden to Russia, to the area where Mordvas live. Those populations living in eastern and western areas have however different admixtures, Mordvas lack Baltic middle-neolithic ancestry and show instead late East European Steppe -like admixture, Sarmatians suggesting incursions of the Iranian speakers. What is also remarkable is that the best Swedish result doesn't include Scania_IA. This doesn't mean that they have not Iron Age ancestry from Scania and in reality they are really close Scania_IA. It means only that present-day Swedes are slightly different, own later admixtures and lack of certain older Baltic admixtures.
QpAdm results are made using distant ancestral references (the right-file) to get best possible coverage. While these results are very reasonable, it is possible to fine-tune all results by using carefully selected less distant references. So these results are directional.
All Finnish groups are from the 1000-genomes project and cover full 1MSNP's, as well as all other groups on the second and third plots. I have seen in my work that testing ancient and modern samples calls for equal coverage of all modern sample groups.
What kind of Fennoscandinavian was the RISE174/Scania_IA? Definitely she was not like a present-day Swede, we can see it in following f3-result below and on PCA-plots.
Next here is a simple PCA-plot, somewhat imperfect one and flat due to a high amount of Asian influence and too few European samples. This plot shows a new Finnish group, Finnx, and binds it to the Finnish ancestry. Finnx forms an ordinary Finnish group with less present-day Swedish admixture than typical Southwest Finns, but also less genetic drift than typical East Finns.
F3-test showing common genetic drift with Scania_IA:
First some European groups in a demonstrative purpose.
South Italians
Chisq and tail prob: 0.436 0.932651
Populations: Beaker_Central_Europe, Iran_LN, Levant_BA
best coefficients: 0.299 0.169 0.531
Jackknife mean: 0.299685849 0.169473438 0.530840713
std. errors: 0.028 0.032 0.033
North Italians
Chisq and tail prob: 1.738 0.628609
Populations: Beaker_Central_Europe, Levant_BA, Iran_LN
best coefficients: 0.542 0.416 0.042
Jackknife mean: 0.542422483 0.415568791 0.042008725
std. errors: 0.019 0.020 0.020
Basques
Chisq and tail prob: 1.789 0.774528
Populations: Beaker_Central_Europe, Iberia_EN
best coefficients: 0.578 0.422
Jackknife mean: 0.579234989 0.420765011
std. errors: 0.030 0.030
Latvians
Chisq and tail prob: 1.101 0.77684
Populations: Scania_IA, Latvia_MN, Evenk
best coefficients: 0.920 0.092 -0.012
Jackknife mean: 0.918423990 0.093588254 -0.012012243
std. errors: 0.042 0.042 0.016
Poles
Chisq and tail prob: 1.699 0.790857
Populations: Poltavka, Germany_MN
best coefficients: 0.502 0.498
Jackknife mean: 0.502425062 0.497574938
std. errors: 0.039 0.039
Estonians
Chisq and tail prob: 0.642 0.886697
Populations: Scania_IA, Latvia_MN, Evenk
best coefficients: 0.933 0.052 0.015
Jackknife mean: 0.931374297 0.053427705 0.015197998
std. errors: 0.039 0.038 0.015
Mordvas
Chisq and tail prob: 0.503 0.973198
Populations: Sarmatian, Scania_IA
best coefficients: 0.472 0.528
Jackknife mean: 0.475543261 0.524456739
std. errors: 0.079 0.079
Swedes
Chisq and tail prob: 1.003 0.9093
Populations: Latvia_LN, England_N
best coefficients: 0.523 0.477
Jackknife mean: 0.520472837 0.479527163
std. errors: 0.052 0.052
Finnx
Chisq and tail prob: 1.647 0.648853
Populations: Scania_IA, Latvia_MN, Evenk
best coefficients: 0.851 0.084 0.065
Jackknife mean: 0.849474735 0.085348593 0.065176672
std. errors: 0.037 0.036 0.014
Some "what if" analyses:
What if Mordvas are Polish
Chisq and tail prob: 11.941 0.0177937
Populations: Poltavka, Germany_MN
best coefficients: 0.727 0.273
Jackknife mean: 0.724786064 0.275213936
std. errors: 0.047 0.047
What if Latvians are Polish
Chisq and tail prob: 9.038 0.0601576
Populations: Poltavka, Germany_MN
best coefficients: 0.563 0.437
Jackknife mean: 0.562264229 0.437735771
std. errors: 0.049 0.049
What if Finnx's are Mordva
Chisq and tail prob: 4.728 0.316351
Populations: Sarmatian, Scania_IA
best coefficients: 0.369 0.631
Jackknife mean: 0.377367376 0.622632624
std. errors: 0.093 0.093
What if East Finns are Mordvas
Chisq and tail prob: 6.593 0.159
Populations: Sarmatian, Scania_IA
best coefficients: 0.317 0.683
Jackknife mean: 0.329388055 0.670611945
std. errors: 0.118 0.118
What if Swedes are Baltic
Chisq and tail prob: 1.867 0.867159
Population: Scania_IA, Latvia_MN, Evenk
best coefficients, Jackknife optimisation is negative, 1.000 0.000 -0.000
The results above show the continuum of Scania_IA from Southern Sweden to Russia, to the area where Mordvas live. Those populations living in eastern and western areas have however different admixtures, Mordvas lack Baltic middle-neolithic ancestry and show instead late East European Steppe -like admixture, Sarmatians suggesting incursions of the Iranian speakers. What is also remarkable is that the best Swedish result doesn't include Scania_IA. This doesn't mean that they have not Iron Age ancestry from Scania and in reality they are really close Scania_IA. It means only that present-day Swedes are slightly different, own later admixtures and lack of certain older Baltic admixtures.