Fortunately, Russian researchers, unlike their Finnish colleagues, always publish the data related to their research, and this way it was possible to download it for my own tests. First, I ran a PCA image on European populations. The picture does not so much aim to tell the position of the Eastern Baltic-Finnic populations on the genetic map of Europe, as to ensure the correctness of the data, excluding possible duplicates and outliers. The picture tells the fact that some new published samples show Baltic-Finnic affinity and identify totally with the Finnish samples well. Those groups are widely known to have a Finnish origin, which was already stated in the study. So there is nothing surprising. Some other new samples in the study didn't show signigicant link with Finns and grouped with Russians, but, as we see in Dstat results, also they are related with Finns. It has to be understood that a many-to-many test like PCA, selecting common genome components of all samples, makes compromises in one-to-one relations. Of particular interest is that the Mordva, who belong to another eastern Finnic group, clearly stand out from the Baltic-Finnic groups and appear to be an approximate Russian-Chuvash mix, while the Baltic-Finnic groups, as their name suggests, place between the Baltic people and the more boreal Finno-Ugric.
It's also interesting to note that my own location is moving more and more towards the Balts. Eastern Baltic-Finnic people don't like me, even though commercial American tests define me as 100% Finnish. This commercial test result contradicts all scientific admixture and PCA analyses, though. This can be explained by the fact that American commercial tests are aimed at Americans who are interested in the familiar matter of where on the old continent their ancestors came from. It is easy to make a dedicated test to identify a population with distinct and local recent (let's say 8-10 generations) ancestry and disregard minor or older genetic drift. Of course this is a dissatisfactory situation for us living on the old continent. This paragraph as a side note.
The material, other than study samples, was downloaded from the Reichlab website without changing anything, except for the Finnish ones, which are from the 1000genomes project. Regarding PCA, I limited the number of Finns to ten random samples. The Dstat data includes all 99 Finns.
Dstat results show strong Finnish kinship in all Karelians and Ingerians, as well as in Veps and Votes. By comparing Finns and Estonians in relation to the east Baltic-Finnic groups of the study, it can be concluded that the Karelians, Ingrians and Veps are most closely related to the Finns and the Estonians to the other east Baltic-Finnic groups of the study. From the comparison between Finns and Russians, it can be concluded that the Novgorod, Pskov and Yaroslavl groups are clearly leaning away from the Finns, but whether this is due to the Estonian or Russian proximity of these research groups remains to be determined. Maybe I'll come back to this later.
Test format: dstat (Finnish,group to compare:study group,outgroup)
Edit 28.12.24 22:00
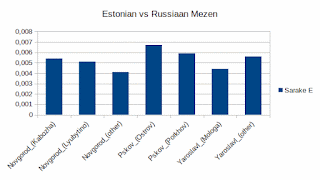
It turned out that the Estonians are cousins of the Baltic-Finnic groups of Novgorod, Pskov and Yaroslavl. Only Andreapol ranks at the same distance as the Estonians when measured by genetic drift.
And I couldn't resist comparing myself to Estonians. In the groups Ingrians, Karelians (Livvi), Karelians (Tver), Novgorod (others) and Yaroslavl (Mologa) we are on the same level (approximately).
Reading about the lost tribes
Mologa history -> link
Novgorod history -> link