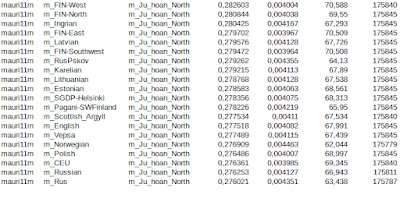
Sorry, I had to continue from what we saw in the study in my previous message. There was two f4-statistics evaluating the origin of populations. It was misleading. We simply can't determine origins by measuring the amount of one ancestral component, not by absolute values or by comparing to another population, because other admixtures has effect on other individual admixtures. We can only measure ratios. Let's assume that a Mesolithic population was 60% EHG and 40% WHG. Then a new migration gave it 50% Steppe admixture. The result after the process was 50% Steppe, 30% EHG and 20% WHG. If we now compare the amount of EHG to any population with different admixing history we get wrong results. I made a statistics showin EHG/WHG ratio of certain populations. I use f3-statistics pop,WHG, Mbuti and pop,EHG,Mbuti. I can't say that it gives absolutely right answers, but f3 is a well-known academic method. EHG and WHG references can even overlap to some extent. In any case, despite of the method the idea using the amount of one admixture proportion to evaluate origin of population is wrong whether it uses absolute or relative values in comparison to another population.
Tuesday, December 20, 2022
Monday, December 19, 2022
A new Finnish study proves slavification in Middle Age Russia
It is like we didn't already know that Uralic speakers in the Russian heartland were Slavicized and replaced by Slavic people. The study found that this happened a few hundreds kilometers from Moscow in the western Oka river area. This is one thing we have known at least 150 years. But what we don't yet know and is more interesting. Why are Baltic Finns, Estonians and Finns, genetically much more western than those people in the supposed Oka river urheimat of Baltic Finnic speakers, and we have been more western back at least 2000 years from now. I can see an explanation in Estonia due to the high Slavic-like R1a concentration, but in Finland we have not such explanation. We have the highest N1c level in Northern Europe and our I1 differs remarkable from all Scandinavian I1 lineages. Common lineages for Finnish and Scaninavian I1 lived 3000 years ago. Yet, we have lived 2000 years in the neighborhood of Saami people, who are in a way semi-Siberians and own 20% Siberian admixture. Without Saami admixture in Finns we would likely be genetically more western than Estonians, regardless our y-dna haplogroups. Estonians got the minuscule Siberian admixture from Finns, especially during the time the Swedish realm reached the Baltic area. How do I know this? Because I have studied genealogy, read Finnish, Estonian and Scandinavian history and migrations between these countries. So what is the problem? It is lack of research of our own genetic background. Looking at the lack of enthusiasm we have in investigating our own history I suggest that we need foreign help in this matter.
A PCA plot from the study with my comment, "Estonian", "Finnish" and "supposed Balt. Finnic urheimat".
Wednesday, December 7, 2022
Who were east Vikings?
East Vikings had influence over wide areas in Baltic and pre-Russian regions. They had permanent strongholds in the area of present day Latvia and on the shores of Ladogan (Staraya Ladoga). Their east ways went through the Bay of Finland and Daugava river in Latvia, which was known earlier as Livonia (known also Dvina river and in Baltic Finnish languages Väinä-joki). Russian chronicles mention them as founders of the earliest Russian state. Some sources connect them to the Byzantine guard known as Varangians, but this information is contradictory. Anyway, it is shown that ancient military and traders from Scandinavia used to make long trips in a wide eastern area.
During those days it was difficult to make difference between aims of Viking trips. They can have been travellers, raiders, traders or even new settlers, or everything this at the same time. But who were they? New ancient samples give some answers. Almost 500 Viking era samples were publishes a few years ago. Were the a homogeneous group? Were they like present-day Scandinavians? My answer is yes, but also no, if we look at genetic picture of all southern Viking Age Swedes.
I made qpDstat tests for samples classified as Vikings, gathered from Sweden and the Baltic area. Mostly they were similar to present-day Scandinavians, but there are also exceptions. I found that samples from Sweden Upsala, Sweden Sigtuna and Gotland were exceptions. Those three had influence from the eastern seaside of the Baltic Sea. On the other hand samples from the Ladogan region and Viking remains found from Saaremaa (in western Estonia) were unambiguously of Scandinavian origin. My conclusion is that Upsala, Sigtuna and Gotland were international trading centres, while eastern travellers were typical Scandinavians and probably from the Southern Scandinavia. The heartland of Viking raiders was in Southern Scandinavia.
Dstat showing how modern Swedes match with Viking Age Scandinavian samples. Positive values mean that modern Swedes were closer to Viking Age Scandinavians than populations after ">"-mark were from the same Viking Age Scandinavians.
Dstat showing geographical Viking Age populations least similar to modern Swedes. Positive values show non-Scandinavian populations being closer to the Viking Age Scandinavians in title than modern Swedes are from the same Viking Age Scandinavian group.
Mauri is me. I have now a sample set fitting with the Human Origins data set and I can do high quality tests with it.
Saturday, December 3, 2022
Who am I?
I have never told here my blog who I am. First, I have a genealogy research based on church and admisistrative documents reaching all my ancestrors to the end of the 17th century and certain Finnish male lineages to the beginning of the 16th century. 99% of my earliest lineages carry Finnish names and were settlers from Tavastia to the seaside of the Bothnian Bay. They supposedly moved from tavastia 500-700 ybp. During the heyday of the Swedish realm, in the 17th century I have some foreign lineages from Baltic countries, Germany and Scotland covering around 3% of my ancestry. From the end of the 18th century my ancestry is again fully Finnish by name, as well as proven by church registers. All this is essential to know before estimating my genetic ancestry. During years I have tested myself on online genetic tests, not only one or two, but maybe tens and I have a good general view about methods and about results I get averagely. Nowadays tests and the research behind them have improved, mostly the improvement is due to the growing amount of reference samples. In a few years the reference sample size is multiplied. However used methods can mislead. Basically methods can be divided into two categories: tests trying to find a prevalent ancestry using genetic fingerprints characteristic for each populations. This method is useful in a worldwide genealogical research, giving a good estimate where your ancestor comes from. Another way to estimate someone's ancestry is an orthodox admixture method using reference groups. The biggest problem in this method is how to select reference groups. Missed or wrong reference groups give false admixtures, because you can practically choose any two or more points on the genetic geography to determine one's admixtures. So be cautious! Nevetheless, real (orthodox) admixture test with wide genome sample can reveal real things with better certainty than the method searching only prevalent ancestry, simply because you typically don't know the used method and how your prevalent ancestry is found and if there are minor admixtures. Both methods can be misleading, but in the case of real admixture you can evaluate things being wrong and right, in the case of prevalent ancestry method you have not data and tools to judge the result.
My first result show that I am Finnish and this data could be published without further information. In this case it tells that my admixture looks MOSTLY like an average Finnish admixture, but some tests could find my Finnish origin by certain genetic fingerprints, which is totally a different case. Showing typical Finnish admixture doesn't tell that the Finns ARE a mixed population, only that it can be in some time frame.
Following results show that I am according these tests, as well the Finns averagely, mixed, but there is a probability that the Finnish reference (possible also others references) is poorly selected. Changing relative position on the genetic geography I could be fully Finnish and the chosen Finnish reference mildly mixed in some other way.
Friday, December 2, 2022
A new Finnish study found last Hunter-Gatheters in Europe
This study claims that Finnish researchers have probably found a hunter-gatheter population that lived only 400 years ago on the northern seaside of the Baltic Sea. Maybe they found the first evidence of a Paleo Diet eater. In the future this kind of random evidences are growing in human remains found on southern seasides in Finland, proving possibly of living hunter-gatherer population that lived there around 2000 AD.
Wednesday, October 5, 2022
Exciting East Hunter-Gatherer ancestry
East Hunter-Gatherer ancestry breaks common logic. While it was in Europe highest in ancient Steppe population, Yamnaya and Afanasievo and next highest in Cored Ware populations, it seems to fade out in modern Baltic populations, although those modern populations have highest Steppe ancestry. Instead of East HG the Balts have highest West HG among all European populations. This made me wonder whether the Dstat, and other tests basing on genetic drift, works correctly. I made a Dstat-run
Dstat(EHG,WHG,<test pop>,Mbuti)
and the results is
To see if there is shift between East and West Hunter-Gatherer drifts I made another test using 3Pop-test, which is rather a test of common ancestries than a comparison. It supports Dstat and shows that the East HG is dominant only in ancient Steppe populations. We can see minor shift though. For example FIN-West shows higher West HG than expected by Dstat. Looking at the time span from the Baltic Corded Ware to the Baltic Bronze Age there is a huge shift from EHG to WHG.
Sunday, August 28, 2022
Common ancestry using heterozygous data
I got an idea to extract heterozygotes from the source population data to see recent admixtures without interference of homozygous inheritance. The goal was to remove local genetic drift and see the recent admixture between population pairs. Several studies have shown that heterozygotes imply recent admixture. It is easy to see that mixing of two or more populations lead to increasing heterozygosity, even homozygous and drifted populations create heterozygosity in mixing process. I removed all homozygotes only from source populations, leaving other populations original to give a chance to homozygous mixtures to lead to heterozygous offsprings. Test samples are from 1000 Genomes- and SGDP-projects modified to fit to the HO-data space published by Reich Lab and in the final stage pruned to include 180000 heterozygous locations, which are then compared to unpruned samples (it is essential to standardize samples in using Admixtools/qp-softwares). Outside listed result I tested also Basque samples. Basque results showed only distant admixture and surprisingly (at least to me) the closest admixture was with British samples, not with IBS and French samples. Again I noticed that Swedish SGDP-samples are useless, which annoy me especially because Sweden belongs to root populations around the Baltic Sea. I would be grateful if someone of my readers has an idea where to download decent Swedish samples fitting with the Human Origin project. I need the HO-space or more to get reasonable amount of heterozygotes.
Mauri11m is me and I was born on the western seaside.
These tests lack of many populations, are not perfect and can only prove the applicability of this method reducing the impact of local genetic drifts.
Tuesday, May 24, 2022
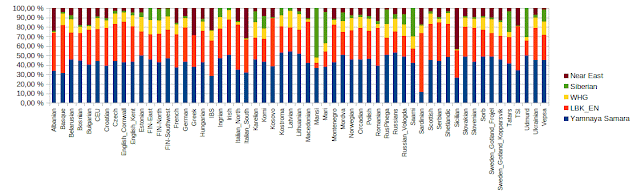
European admixtures by qpAdm
What I have seen we have no standards in reporting ancient admixtures of recent populations. Commercial services give sometimes 70% Western Hunter Gatherer, sometimes 15%. Methods are poorly described, if at all. Obviously there are admixture overlaps and the results depend on how admixture tools identify them. For instance Yamnaya Samara overlaps with several hunter gatherer groups. I am sure that also European early farmers (LBK_EN) overlap with later Near East admixture in Europe and later Near Eastern admixture exists in Mediterranean. In other words admixture runs without Near Eastern samples assume EEF, but it doesn't give best fit. My test uses following qpAdm files:
Right file:
Mbuti
Levant_Neolithic (sample I0867)
Onge
Iran_N
Villabruna
Mixe
Ami
Krasnoyarsk_Siberian
Itelmen
Left file:
source
Yamnaya_Samara
LBK_EN
WHG
Nganasan
BedouinB (selected most distant samples from Europeans)
The results looks reasonable and qpAdm gave positive best coefficients without errors with only one exception, Swedes. I don't know why modern Swedes failed, because ancient Gotlanders made a perfect fit.
Tuesday, May 17, 2022
New Finnish Iron Age study on the way
Here the link of preprint. It looks like an arrival of a new male group from Estonia representing Baltic Finns mixed with older more Swedish-like population. Obviously those western females represented older local population, which at the end of the Viking Era became isolated from Sweden. Hard to believe that Swedish females migrated to Finland without men.
Wednesday, April 13, 2022
New blog
Unfortunately it looks like Finnish genetic research has been paused so far due to disappointing results. Hopefully Finnish researchers make necessary reappraisals, because we need objective knowledge about our prehistory. Today the gap between linguistics, genetics and archaeology is too big to support objectives of Finnish genetic research. I am afraid it takes years to turn the ship into a correct lane and I probably never will see rational Finnish genetic research. My pessimism is based on that fact that I have over 10 years experience about this matter.
Meanwhile I can do nothing with this matter I decided to open a new blog dealing with Finnish history and prehistory. Topics will mainly cover observations mirroring the Western Finnish history, facts, myths and visions. Unfortunately I have not time and resources to give inclusive reference lists and readers have to make some efforts for it. Language is often Finnish, but translators are today reasonable good to give you points.
Monday, January 10, 2022
Kazakh ancient N-haplogroups were N1a1a1a1a
Several samples from the time around 300 BC in Kazakhstan belong to the same haplogroup level than Kola Peninsula samples from 1500 BC, reported by mutations L1026 or L392. I was not able to find any RECENT ISOGG downstream mutations, which makes me think if all European side N can be derived from these Kazakh Iron Age men. If this is true we have a problem with the home land of Finnic speakers, considering that they belonged to N and home land of them was not near Kazakhstan. At 500 BC we should find downstream mutation taking into account datings of the first Finnic speakers in the Baltic area and believing in the incidence of N1a1a1a1a at 500 BC in Europe and in Central Asia simultaneously. TMRCA of the N1a1a1a1a is according to Yfull 3000 years (1000 BC). The mystery of the origin of European N remains. Everything proves about a rapid expansion to the Baltic area and to North Russia. What ever caused this, what language they spoke and adapted remains without answer and makes the mystery even deeper after these finds from Kazakhstan. Another explanation is that there was still unknown brance of N1a1a1a1a somewhere in West Siberia or Northern Russia around 1000-500 BC, but we have not evidence of it.
SMV001 N1a1a1a1a Sargat_300BCE
BIY001 N1a* Sargat_300BCE
BIY002 N1a1a1a1a Sargat_300BCE
BIY005 N1a1a1a1a Sargat_300BCE
BIY007 N1a1a1a1a Sargat_300BCE
BIY009 N1a1a1a1a Sargat_300BCE
BIY012 N* Sargat_300BCE
KOK001 N1a* (likely N1a1a1a1a) Kokonovka_200BCE
KOK002 N1a1a1a1a Kokonovka_200BCE
BGD004 N1* (likely N1a1a1a1a) Bogdanovka_150BCE
https://www.science.org/doi/10.1126/sciadv.abe4414
N1a1a1a1a L392 L1026