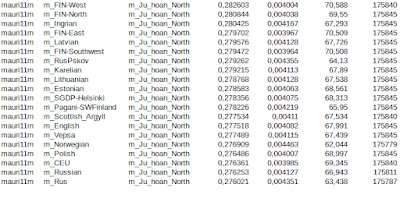
I got an idea to extract heterozygotes from the source population data to see recent admixtures without interference of homozygous inheritance. The goal was to remove local genetic drift and see the recent admixture between population pairs. Several studies have shown that heterozygotes imply recent admixture. It is easy to see that mixing of two or more populations lead to increasing heterozygosity, even homozygous and drifted populations create heterozygosity in mixing process. I removed all homozygotes only from source populations, leaving other populations original to give a chance to homozygous mixtures to lead to heterozygous offsprings. Test samples are from 1000 Genomes- and SGDP-projects modified to fit to the HO-data space published by Reich Lab and in the final stage pruned to include 180000 heterozygous locations, which are then compared to unpruned samples (it is essential to standardize samples in using Admixtools/qp-softwares). Outside listed result I tested also Basque samples. Basque results showed only distant admixture and surprisingly (at least to me) the closest admixture was with British samples, not with IBS and French samples. Again I noticed that Swedish SGDP-samples are useless, which annoy me especially because Sweden belongs to root populations around the Baltic Sea. I would be grateful if someone of my readers has an idea where to download decent Swedish samples fitting with the Human Origin project. I need the HO-space or more to get reasonable amount of heterozygotes.
Mauri11m is me and I was born on the western seaside.
These tests lack of many populations, are not perfect and can only prove the applicability of this method reducing the impact of local genetic drifts.







No comments:
Post a Comment
English preferred, because readers are international.
No more Anonymous posts.