Here, as a showcase of the new data two PCA prints and some comments. Instead of making a central continental European picture I included four outgroups to see the effect. Those outgroups are Armenians, Mongolians, Sardinians and Saamis. For the present individual names are picked straight from original sources and can be somewhat ambiguous.
As we can see we have several clusters, which makes possible to evaluate the data. For example Scandinavians of SGDP and Pagani et al. cluster with East Europeans.
Personally I don't give much attention to PCA-figures, because the result depends on the selected samples, amounts, ratios between populations sizes, about how mixed are individuals etc. My upcoming high resolution tests will be much more interesting.
Added time 12:50
If someone is interested in how Mordvas locate on this map. They are very similar to North Russians and RusKU (Pagani et al.) and move towards Mongolians. Baltic Finns moves towards Saamis. Sorry, GIMP makes something unwanted with colors.
I



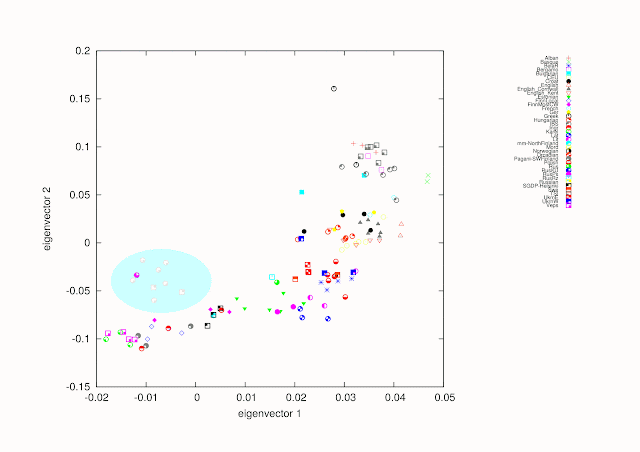
The Greek sample clustering with Armenians must be from an Anatolian Greek (most probably from an eastern Pontian Greek giving its strong clustering with Armenians). This explains why it is not in the South European genetic cluster.
ReplyDeleteNote: I have seen many Anatolian Greek genetic results and know well how genetically different they are from Balkan Greeks.
Very possible, these high resolution samples are quite diverse and in many cases atypical. Another example Pagani's Finns actually live in Estonia, are likely Estonian citizens and they don't give exact ancestral information. Instead of this SGDP's samples are likely from 1000genomes, newly scanned of course. Pretty good work. Pagani's decision is inexplicable, like searching Brits from America instead of using already available British samples. They also give wrong locations (geographic coordinates).
Delete