It was worth of waiting for a few weeks to see these Irish samples, especially because I already expected that Irish insular samples could reveal new things about ancient people who lived in Northwest Europe. You see the original study here. There are four samples, three from Rathlin Island in Northern Ireland and one sample from Ballynahatty, which locates in Northern Ireland. Two of Rathlin samples are of low quality and don't work well with my database based on Estonian Biocentre's data. Maybe I'll download them later to the Lazaridis' database. The third Rathlin and Ballynahatty samples are however excellent.
Picking from the study
- Ballynahatty, a Neolithic woman (3343–3020 cal BC)
- Rathlin, in context of an early megalithic passage-like grave, an Early Bronze Age man from Rathlin Island (2026–1885 cal BC)
I was really excited when started to analyse Rathlin samples, because it was possible that it would reveal new knowledge about ancient people who lived in North Europe before eastern Bronze Age steppe migrations. I decided to compare them to present-day population instead of using ancient samples, to make results touchable. At first I tested which of modern populations are closest Rathlin and Ballynahatty samples and found that the Rathlin genome emphasized still Irish people. Ballynahatty sample was closest present-day Sardinians, representing typical Neolithic era.
After processing all this from fastq-files 1) I made two qpDstat comparisons to find out who of modern populations resembles best those ancient Irish samples in comparison with best fits of modern populations. In comparison with the Rathlin man I included also my project samples, mainly Finnish and Swedish individuals.
Rathlin and modern populations
Ballynahatty and modern populations
Inspired by the western origin of Saami people I made one comparison more using another database to get reliable results with the Saami sample introduced by Haak et al. 2015. It looks like, despite of the remarkable North Asian admixture, they have Rathlin like ancestry more than Eastern Finns, who have less North Siberian.
Saami between ancient samples, using the arrangement seen already in my previous post
FI15 is from Northern Karelia, FI12 is western Finnish, FI10 is from Finnish Lapland.
Finally, after downloading and testing DNA.LAND's admixture program, I made some admixture analyses. You can find and download the software from their site, here. This small program is based on allele frequencies and probably the method is Markov chain Monte Carlo. It is not based on original alleles and genetic drift, thus there is always a residual admixture. There are also other weaknesses, what kind of, it could be a new topic. Now I only say that in my opinion it has problems in composing kinship populations with different minor admixtures.
Two results using references downloaded from DNA.LAND
Rathlin1
CSAMERICA 0.00697236
KALASH 0.0165295
NEEUROPE 0.223957
NEUROPE 0.731415
PATHAN-SINDHI-BURUSHO 0.00252671
SWEUROPE 0.0185991
Ballynahatty
ITALY 0.0116662
SARDINIA 0.565326
SWEUROPE 0.423008
Two results using my Estonian-BC database as reference
Rathlin1
Bulgaria 0.0524726
Colombian 0.00995061
Ireland 0.213786
Kalash 0.00510618
Latvia 0.0102334
Lithuania 0.221711
Orcadian 0.140857
RU_Smolensk 0.0244413
Scotland 0.207837
Udmurtia 0.0262457
Welsh 0.0840646
Ballynahatty
Basque 0.0446958
Ireland 0.0547766
NorthItaly 0.0953578
Sardinian 0.569061
Scotland 0.0409351
Sicily 0.0670495
Spain 0.12727
Tuscany 0.000854723
1) I have changed my fastq-process. Although BWA is an excellent program in mapping reads, it's automatic trimming is not powerful enough and now I have rerun also all older samples using separate trimming program.


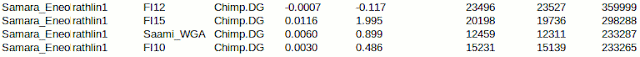
No comments:
Post a Comment
English preferred, because readers are international.
No more Anonymous posts.